Resazurin Assay Kit (Cell Viability) (ab129732)
Key features and details
- Assay type: Cell-based (quantitative)
- Detection method: Fluorescent
- Platform: Microplate reader
- Sample type: Adherent cells, Suspension cells
Overview
-
Product name
Resazurin Assay Kit (Cell Viability)
See all Cell viability/proliferation kits -
Detection method
Fluorescent -
Sample type
Adherent cells, Suspension cells -
Assay type
Cell-based (quantitative) -
Product overview
Resazurin Assay Kit ab129732 is a fluorometric/colorimetric assay used to measure the metabolic capacity of live cells.
The resazurin assay is commonly used to quantify the number of live cells in a sample, and to monitor cell viability / cytotoxicity. It is a fast, simple, accurate and homogeneous / no-wash high throughput assay that can be used to monitor cells for up to 24-48 hours.
The resazurin assay protocol is based on the reduction of oxidized non-fluorescent blue resazurin to a red fluorescent dye (resorufin) by the mitochondrial respiratory chain in live cells. The amount of resorufin produced is directly proportional to the number of living cells. Resorufin has Ex/Em of 530-560 / 590 nm and absorbance at absorbance at 570nm.
The resazurin dye can be measured in fluorescence or absorbance mode. However fluorescence mode measurement offers greater assay linearity, reproducibility, robustness and sensitivity.
Resazurin assay protocol summary:
- add resazurin reagent to cells in growth media and incubate for 4 hr at 37ºC
- analyze with microplate reader -
Notes
This kit was previously called Mitochondrial Viability Stain.
Review our cell health assay guide to learn about our kits to perform a cell viability assay, cytotoxicity assay or cell proliferation assay.
-
Platform
Microplate reader
Properties
-
Storage instructions
Store at +4°C. Please refer to protocols. -
Components 2000 tests 20X Cell Viability Stain 1 x 24ml -
Research areas
-
Relevance
Cell proliferation is the multiplication or reproduction of cells, as a result of cell growth and cell division, resulting in the expansion of a cell population. -
Alternative names
- Cell Cytotoxicity
- cell tracking
Images
-
Jurkat cells were seeded at 25,000 cells per well in a 50 µL volume and immediately overlayed with a 2X concentration of Staurosporin for a final volume of 100 µL. Cells were incubated for 4 hours with Staurosporin prior to the addition of 2X stain diluted in RPMI media. After 4 hours of further incubation with stain, fluorescence was measured. IC50 for staurosporin was found at 500 nM.
-
Jurkat cells were seeded at 25,000 cells per well in a 50 µL volume and immediately overlayed with a 2X concentration of the indicated drug for a final volume of 100 µL. Cells were incubated for 2 hours with the indicated drug prior to the addition of 2X stain diluted in RPMI media. After 4 hours of further incubation with stain, fluorescence was measured. IC50 for the indicated drug was found at 22 – 25 µM.
-
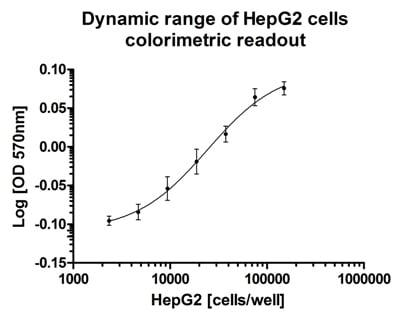
Dynamic range and suitability of the mitochondrial viability assay for high throughput screening. Cells were seeded in a titration series with n=11 for each data point. Colorimetric readout gave on average a Z factor of 0.72 on HepG2 cells seeded from 40,000 – 150,000 cells per well.
-
Dynamic range and suitability of the mitochondrial viability assay for high throughput screening. Cells were seeded in a titration series with n=11 for each data point. Fluorometric readout gave on average a Z factor of 0.82 on Jurkat cells seeded from 1,500 – 1000,000 cells per well on a 96-well plate.
-
Mitochondrial viability stain in long term toxicity. HepG2 cells previously cultured in glucose or galactose substrate media were seeded at 10,000 cells per well and allowed to adhere overnight. Media was replaced for the specific culture media in the presence of a titration series of Rotenone (5 µM – 0.5 pM). Cells were treated for 72 hours prior to the addition of 2X stain. After 4 hours of incubation plates were analyzed by fluorescent readout.
-
Effect of stain incubation time on assay reproducibility and background. Dark wall plates were seeded with a titration series of each cell line in a final volume of 100 µL of media. 2X stain (diluted in the cell line specific growth media) was added to each well at specified time points. XY graphs data is shown after media and background substraction. Bar graph shows mean background staining at different time points. 4 hour incubation of stain results in optimal reproducibility with minimal increase of background over the 2 hour incubation.