Chromatin Accessibility Assay Kit (ab185901)
Key features and details
- Assay type: Quantitative
- Assay time: 1 hr 30 min
- Sample type: Adherent cells, Suspension cells, Tissue
Overview
-
Product name
Chromatin Accessibility Assay Kit -
Sample type
Tissue, Adherent cells, Suspension cells -
Assay type
Quantitative -
Assay time
1h 30m -
Species reactivity
Reacts with: Mammals -
Product overview
Chromatin Accessibility Assay Kit ab185901 is a complete set of optimized reagents designed for conducting a gene-specific analysis of chromatin accessibility including nucleosome/transcription factor positioning from various biological samples via real time PCR.
The chromatin accessibility assay works on the principle that accessible chromatin is easily accessed by a nuclease mix to digest the DNA. When the DNA from the nuclease-treated sample and a no-nuclease control is analyzed for a particular gene by PCR, the more accessible the chromatin at that gene, the lower the signal of the nuclease-treated sample relative to the no-nuclease control.
Chromatin accessibility assay protocol summary:
- lyze cells and extract chromatin by adding lysis buffer to cell pellet, resuspending and incubating for 10 min
- centrifuge at 5,000 rpm for 5 min, discard supernatant, resuspend chromatin pellet in wash buffer and centrifuge again
- resuspend chromatin pellet in nuclease reaction mix (or no-nuclease control mix to controls) and incubate for 4 min
- add stop solution and incubate for 10 min
- add Proteinase K and incubate for 60 min
- add samples to DNA binding columns, spin, and discard flow through
- add DNA washing solution to columns, spin, and discard flow through 3 times
- add elution solution and spin to elute DNA
- analyze gene target with PCR and compare nuclease-treated to no-nuclease control -
Notes
The accessibility of regulatory elements in chromatin is critical for many aspects of gene regulation. Nucleosomes positioned over regulatory elements inhibit access of transcription factors to DNA. To elucidate the role of the interactions between chromatin and transcription factors, it is crucial to determine chromatin accessibility through mapping of the nucleosome positioning along the genome. In general, the more condensed the chromatin, the more difficult it is for transcription factors and other DNA binding proteins to access DNA and carry out their tasks. The more accessible the DNA, the more likely surrounding genes are actively transcribed. The presence (or the absence) of nucleosomes directly or indirectly affects a variety of other cellular and metabolic processes such as recombination, replication, centromere formation, and DNA repair.
Properties
-
Storage instructions
Please refer to protocols. -
Components 48 tests 1000X Protease Inhibitor Cocktail 1 x 50µl 10X DNA Washing Solution 1 x 3ml 10X Lysis Buffer 1 x 11ml 10X Wash Buffer 1 x 10ml DNA Binding Solution 1 x 15ml Elution Solution 1 x 1ml F-Collection Tube 1 x 50 units F-Spin Column 1 x 50 units Negative Control Primer-F (20 μM) 1 x 10µl Negative Control Primer-R (20 μM) 1 x 10µl Nuclear Digestion Buffer 1 x 3ml Nuclease Mix 1 x 50µl Positive Control Primer-F (20 μM) 1 x 10µl Positive Control Primer-R (20 μM) 1 x 10µl Proteinase K (10 mg/mL) 1 x 110µl Reaction Stop Solution 1 x 0.5ml -
Research areas
Images
-
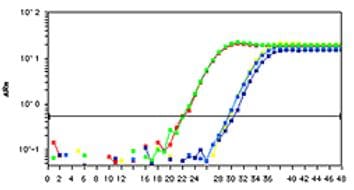
Amplification of proximal promoter regions for the constitutively expressed target gene was carried out in MDA-231 cells by using positive control primers. Red and green lines: Nse-untreated; Blue lines: Nse-treated. The difference of Ct values between Nse-treated and untreated is > 7 cycles.
-
Amplification of proximal promoter regions for the constitutively repressed target gene was carried out in MDA-231 cells by using negative control primers. Red and green lines: Nse-untreated; Blue and pink lines: Nse-treated. The difference of Ct values between Nse-treated and untreated is