RubyGlow™ Bacterial assay (Luminescent) (ab189820)
Key features and details
- Detection method: Luminescent
- Platform: Microplate reader
Overview
-
Product name
RubyGlow™ Bacterial assay (Luminescent) -
Detection method
Luminescent -
Product overview
Bacterial Assay (Luminescent) (ab189820) provides an easy and quick method to detect and quantify bacterial cells based on ATP present in metabolically active cells. The kit contains a genetically engineered luciferase which generates very stable luminescent signals. This luciferase emits red upon luciferin conversion and thus can be multiplexed with other assays using blue or green fluorescence.
-
Notes
The bacterial enumeration in different bacterial samples is of great importance in many fields of research. Traditional methods usually take several days. For fast results, one of the methods to estimate the number of microbial cells is based on the determination of adenosine triphosphate (ATP) levels, assuming that living cells are generating ATP and the ATP content of microbial cells is relatively constant across all phases of growth. One of the most sensitive techniques for measurement of ATP levels has proven to be the luciferin-luciferase bioluminescent assay. When ATP is the limiting component in the reaction, the intensity of the emitted light will be proportional to ATP concentration in the sample.
The kit utilizes a new luciferase that exhibits a long-wavelength light emission (619 nm), as well as improved thermostability, compared to the native green firefly luciferase often used in these assays. This unique feature enables this assay kit to be compatible with assays using standard luciferase analyses, or other green (fluorescein-based) or coumarin (blue color) assay methods.
-
Platform
Microplate reader
Properties
-
Storage instructions
Please refer to protocols. -
Components 1 packs RubyGlow™ Luciferase 1 x 0.5mg Luciferin Substrate 1 x 1.4mg Lysis Buffer 1 x 12ml Lysozyme 1 x 20mg Phosphate/EDTA Buffer 1 x 2ml Reaction Buffer 1 x 6ml
Images
-
Cultures from JM109 or BL21(DE3)pLysS (chloramphenicol resistance) strains harboring an expression vector (confer ampicillin resistance) were diluted and mixed with a number of antibiotics. This mixture was incubated at 37ºC for 5 hours. Then cells were lysed, ATP extracted and measured with ab189820 Bacterial Assay (Luminescent). Luminescence was recorded. RLU was plotted against the different antibiotics. AMP:ampicillin; Tet:tetracyclin; Strep:streptomycin; CAM:chloramphenicol. Ampicillin and chloramphenicol inhibited the growth of JM109, but didn’t affect the growth of resistant strain.
-
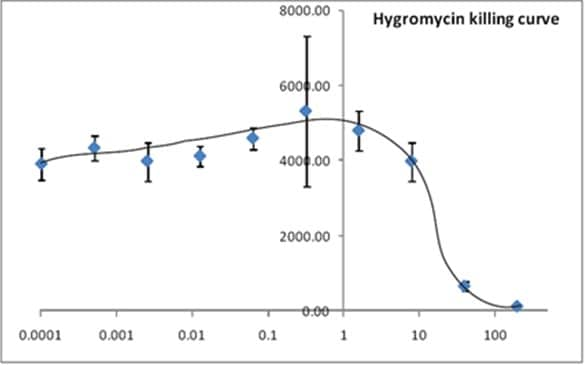
Cultures from JM109 were diluted and mixed with a series of Hygromycin dilutions. This mixture was incubated at 37ºC for 5 hours, lysed, ATP extracted and measured with ab189820 Bacterial Assay (Luminescent). Luminescence was recorded. RLU was plotted against Hygromycin concentration.
-
Dilutions from exponentially grown W3110 were lysed, ATP extracted and luminescence recorded. To examine whether extended incubation at 37ºC will help the detection of high dilution culture, we tried 2 and 4 hour incubation periods and found that the detection limit was dropped at least two magnitudes.
-
Dilutions from exponentially grown W3110 were lysed, ATP extracted and luminescence recorded. RLUs were plotted against the bacterial growth which was indicated by OD600. Linearity was observed up to OD600 of 0.8. The insert shows the linearity below OD600 of 0.1.